Ensemble of Convolutional Neural Networks to diagnose Acute Lymphoblastic Leukemia from microscopic images
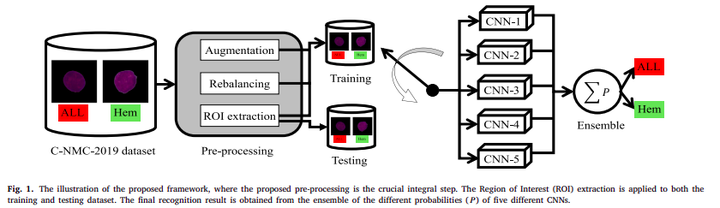 Proposed framework
Proposed framework
Abstract
Acute Lymphoblastic Leukemia (ALL) is a blood cell cancer characterized by the presence of excess immature lymphocytes., Even though automation in ALL prognosis is essential for cancer diagnosis, it remains a challenge due to the morphological correlation between malignant and normal cells. The traditional ALL classification strategy demands that experienced pathologists read cell images carefully, which is arduous, time-consuming, and often hampered by interobserver variation. This article has automated the ALL recognition task by employing deep Convolutional Neural Networks (CNNs). The weighted ensemble of deep CNNs is explored to recommend a better ALL cell classifier. The weights are estimated from ensemble candidates’ corresponding metrics, such as F1-score, area under the curve (AUC), and kappa values. Various data augmentations and pre-processing are incorporated to achieve a better generalization of the network. Our proposed model was trained and evaluated utilizing the C-NMC-2019 ALL dataset. The proposed weighted ensemble model has outputted a weighted F1-score of 89.7%, a balanced accuracy of 88.3%, and an AUC of 0.948 in the preliminary test set. The qualitative results displaying the gradient class activation maps confirm that the introduced model has a concentrated learned region. In contrast, the ensemble candidate models, such as Xception, VGG-16, DenseNet-121, MobileNet, and InceptionResNet-V2, separately exhibit coarse and scatter learned areas in most cases. Since the proposed ensemble yields a better result for the aimed task, it can support clinical decisions to detect ALL patients in an early stage.