Publications
Gestational diabetes mellitus (GDM) is characterized by glucose intolerance during pregnancy, resulting in an elevated blood glucose level and short-term and long-term health burdens. Therefore, early screening would aid in reducing complications associated with GDM and adverse pregnancy outcomes. Machine learning (ML) algorithms are a promising alternative to manual GDM early-stage assessment. In this article, we propose a machine learning (ML) pipeline that employs five distinct classifiers: decision trees (DT), linear discriminant analysis (LDA), logistic regression (LR), XGBoost (XGB), and Gaussian naive Bayes (GNB). Our framework incorporates the essential preprocessing stages, such as filling in missing values, selecting important features, tuning hyperparameters, and applying stratified K-fold cross-validation to improve the model’s robustness and precision. The K-Nearest Neighbors (KNN) method outperforms the other strategies in the proposed framework based on a comprehensive analysis of three distinct missing data imputation techniques. In addition, eight out of fifteen features are chosen, implementing a procedure for feature selection. Finally, when the XGB classifier is combined with the presented preprocessing, the performance improves by significant margins, yielding the utmost achievable accuracy of 0.9719 and an area under the ROC curve of 0.9982. This promising result makes our pipeline useful for GDM prediction in the earliest stages.
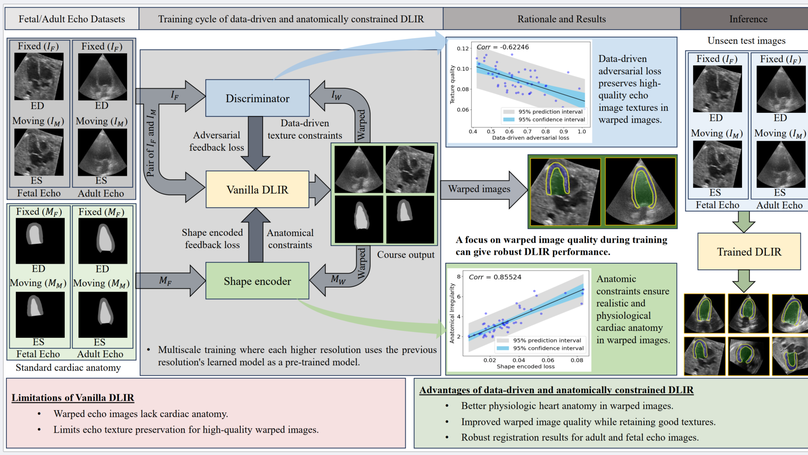
Temporal echocardiography image registration is important for cardiac motion estimation, myocardial strain assessments, and stroke volume quantifications. Deep learning image registration is a promising way to achieve consistent and accurate registration results with low computational time. We propose that a greater focus on the warped image’s anatomic plausibility and image texture can support robust results and show that it has sufficient robustness to be applied to both fetal and adult echocardiography. Our proposed framework includes (1) an anatomic shape-encoded loss to preserve physiological myocardial and left ventricular anatomical topologies, (2) a data-driven loss to preserve good texture features, and (3) a multi-scale training of a data-driven and anatomically constrained algorithm to improve accuracy. Our experiments demonstrate a strong correlation between the shape-encoded loss and good anatomical topology and between the data-driven loss and image textures. They improve different aspects of registration results in a non-overlapping way. We demonstrate that these methods can successfully register both adult and fetal echocardiography using the public CAMUS adult dataset and our fetal dataset, despite the inherent differences between adult and fetal echocardiography. Our approach also outperforms traditional non-DL gold standard registration approaches, including optical flow and Elastix, and could be translated into more accurate and precise clinical quantification of cardiac ejection fraction, demonstrating potential for clinical utility.
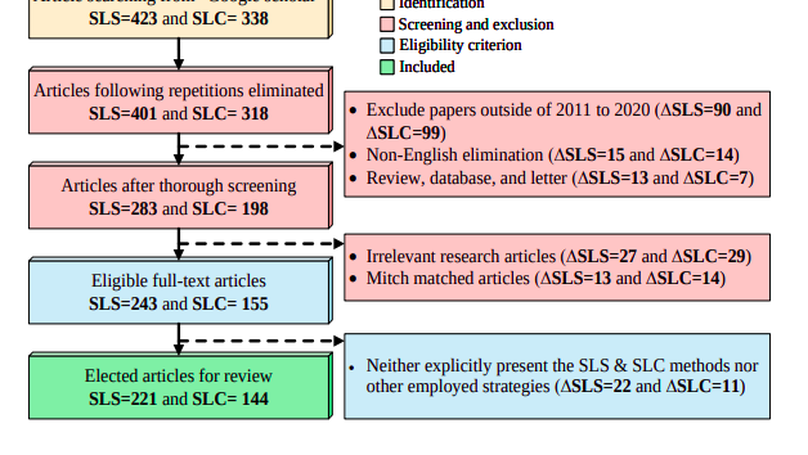
The Computer-aided Diagnosis (CAD) system for skin lesion analysis is an emerging field of research that has the potential to relieve the burden and cost of skin cancer screening. Researchers have recently indicated increasing interest in developing such CAD systems, with the intention of providing a user-friendly tool to dermatologists in order to reduce the challenges that are raised by manual inspection. The purpose of this article is to provide a complete literature review of cutting-edge CAD techniques published between 2011 and 2020. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) method was used to identify a total of 365 publications, 221 for skin lesion segmentation and 144 for skin lesion classification. These articles are analyzed and summarized in a number of different ways so that we can contribute vital information about the methods for the evolution of CAD systems. These ways include: relevant and essential definitions and theories, input data (datasets utilization, preprocessing, augmentations, and fixing imbalance problems), method configuration (techniques, architectures, module frameworks, and losses), training tactics (hyperparameter settings), and evaluation criteria (metrics). We also intend to investigate a variety of performance-enhancing methods, including ensemble and post-processing. In addition, in this survey, we highlight the primary problems associated with evaluating skin lesion segmentation and classification systems using minimal datasets, as well as the potential solutions to these plights. In conclusion, enlightening findings, recommendations, and trends are discussed for the purpose of future research surveillance in related fields of interest. It is foreseen that it will guide researchers of all levels, from beginners to experts, in the process of developing an automated and robust CAD system for skin lesion analysis.
Breast Cancer (BC) is one of the numerous typical diseases worldwide, occurring in 22.9% of all cancers in women and causing 13.7% of cancer deaths. The BC prognosis is highly demanded to increase the survival rate of the patient suffering from BC. Throughout this paper, an automated decision-making pipeline for BC detection has been proposed, incorporating Machine Learning (ML) algorithms like Gaussian Naive Bayes (GNB), Random Forest (RF), XGBoost (XGB), AdaBoost (AdB), and preprocessing such as Outlier Rejection (OR) and Attribute Selection (AS). A weighted ensemble of ML models has been recommended in the introduced pipeline. The experiments are trained and evaluated using Breast Cancer Wisconsin’s public dataset from UCI Repository, employing five-fold cross-validation. The best possible accuracy obtained from the proposed framework is 97.0%, with the utilization of seventeen features out of a total of thirty. The observed results conclude that a weighted ensemble of AdB and XGB in conjunction with OR and AS as a preprocessing can successfully enhance the BC detection outcomes with a significantly short execution time of 2.10 seconds.
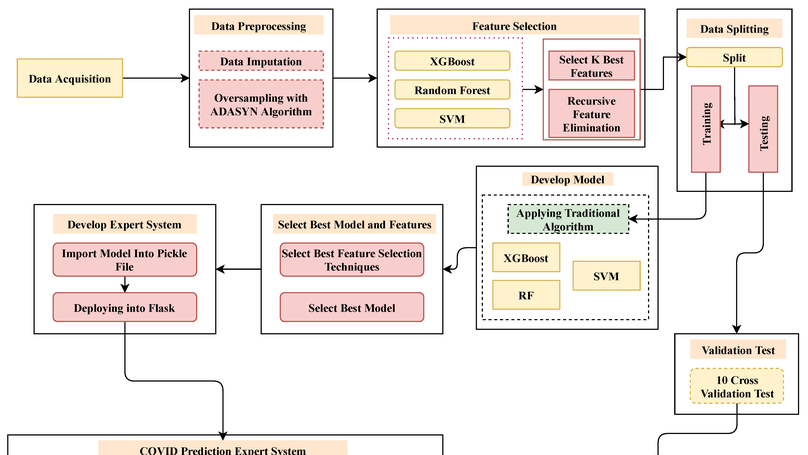
COVID-19 has imposed many challenges and barriers on traditional healthcare systems due to the high risk of being infected by the coronavirus. Modern electronic devices like smartphones with information technology can play an essential role in handling the current pandemic by contributing to different telemedical services. This study has focused on determining the presence of this virus by employing smartphone technology, as it is available to a large number of people. A publicly available COVID-19 dataset consisting of 33 features has been utilized to develop the aimed model, which can be collected from an in-house facility. The chosen dataset has positive and negative samples, demonstrating a high imbalance of class populations. The Adaptive Synthetic (ADASYN) has been applied to overcome the class imbalance problem with imbalanced data. Ten optimal features are chosen from the given 33 features, employing two different feature selection algorithms, such as K Best and recursive feature elimination methods. Mainly, three classification schemes, Random Forest (RF), eXtreme Gradient Boosting (XGB), and Support Vector Machine (SVM), have been applied for the ablation studies, where the accuracy from the XGB, RF, and SVM classifiers achieved , , and , respectively. As the XGB algorithm confers the best results, it has been implemented in designing the Android operating system base and web applications. By analyzing 10 users’ questionnaires, the developed expert system can predict the presence of COVID-19 in the human body of the primary suspect. The preprocessed data and codes are available on the GitHub repository.
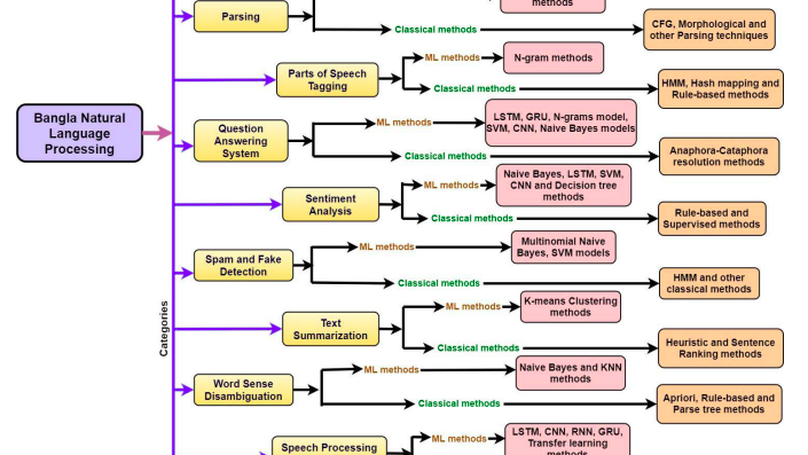
The Bangla language is the seventh most spoken language, with 265 million native and non-native speakers worldwide. However, English is the predominant language for online resources and technical knowledge, journals, and documentation. Consequently, many Bangla-speaking people, who have limited command of English, face hurdles to utilize English resources. To bridge the gap between limited support and increasing demand, researchers conducted many experiments and developed valuable tools and techniques to create and process Bangla language materials. Many efforts are also ongoing to make it easy to use the Bangla language in the online and technical domains. There are some review papers to understand the past, previous, and future Bangla Natural Language Processing (BNLP) trends. The studies are mainly concentrated on the specific domains of BNLP, such as sentiment analysis, speech recognition, optical character recognition, and text summarization. There is an apparent scarcity of resources that contain a comprehensive review of the recent BNLP tools and methods. Therefore, in this paper, we present a thorough analysis of 75 BNLP research papers and categorize them into 11 categories, namely Information Extraction, Machine Translation, Named Entity Recognition, Parsing, Parts of Speech Tagging, Question Answering System, Sentiment Analysis, Spam and Fake Detection, Text Summarization, Word Sense Disambiguation, and Speech Processing and Recognition. We study articles published between 1999 to 2021, and 50% of the papers were published after 2015. Furthermore, we discuss Classical, Machine Learning and Deep Learning approaches with different datasets while addressing the limitations and current and future trends of the BNLP.
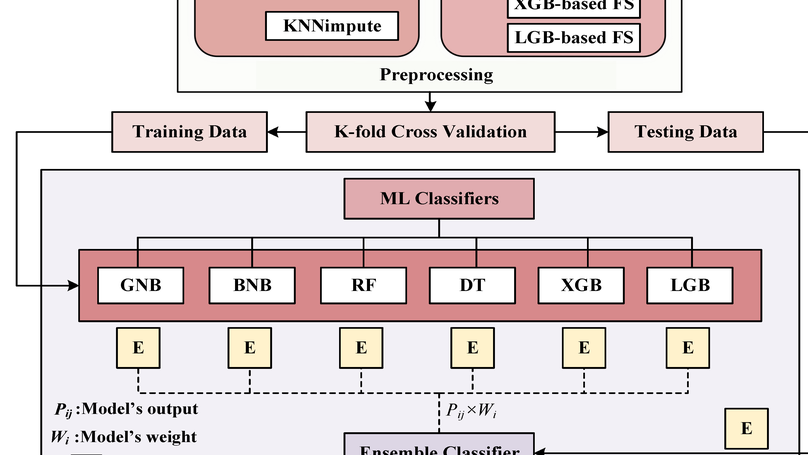
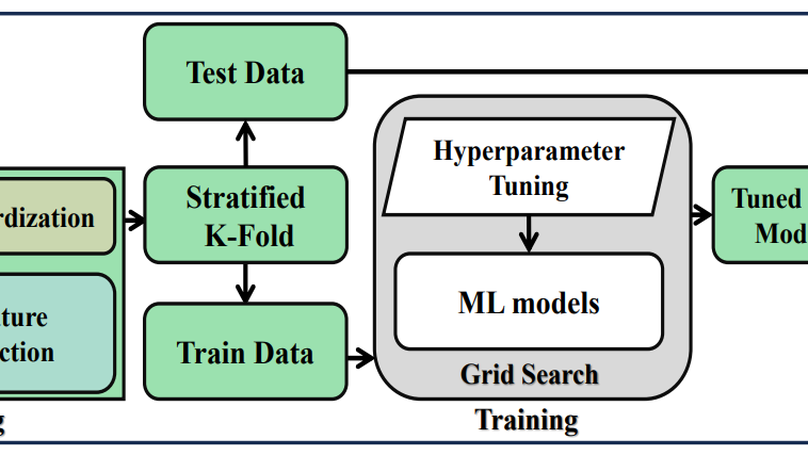
Diabetes is one of the most rapidly spreading diseases in the world, resulting in an array of significant complications, including cardiovascular disease, kidney failure, diabetic retinopathy, and neuropathy, among others, which contribute to an increase in morbidity and mortality rate. If diabetes is diagnosed at an early stage, its severity and underlying risk factors can be significantly reduced. However, there is a shortage of labeled data and the occurrence of outliers or data missingness in clinical datasets that are reliable and effective for diabetes prediction, making it a challenging endeavor. Therefore, we introduce a newly labeled diabetes dataset from a South Asian nation (Bangladesh). In addition, we suggest an automated classification pipeline that includes a weighted ensemble of machine learning (ML) classifiers: Naive Bayes (NB), Random Forest (RF), Decision Tree (DT), XGBoost (XGB), and LightGBM (LGB). Grid search hyperparameter optimization is employed to tune the critical hyperparameters of these ML models. Furthermore, missing value imputation, feature selection, and K-fold cross-validation are included in the framework design. A statistical analysis of variance (ANOVA) test reveals that the performance of diabetes prediction significantly improves when the proposed weighted ensemble (DT + RF + XGB + LGB) is executed with the introduced preprocessing, with the highest accuracy of 0.735 and an area under the ROC curve (AUC) of 0.832. In conjunction with the suggested ensemble model, our statistical imputation and RF-based feature selection techniques produced the best results for early diabetes prediction. Moreover, the presented new dataset will contribute to developing and implementing robust ML models for diabetes prediction utilizing population-level data.
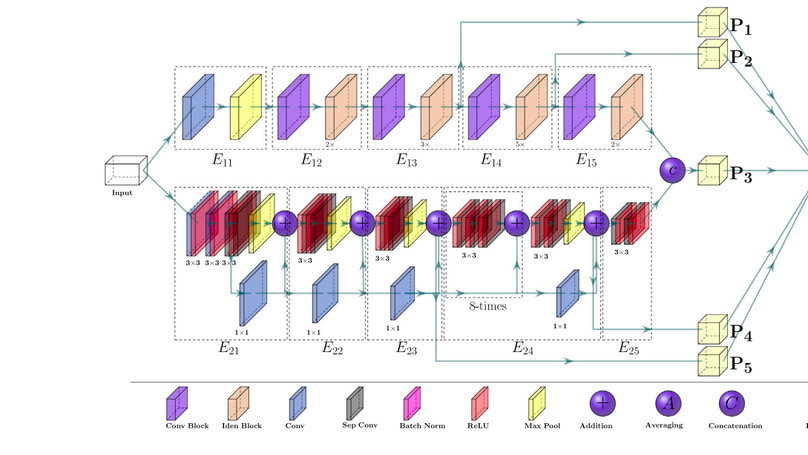
Since the COVID-19 pandemic, several research studies have proposed Deep Learning (DL)-based automated COVID-19 detection, reporting high cross-validation accuracy when classifying COVID-19 patients from normal or other common Pneumonia. Although the reported outcomes are very high in most cases, these results were obtained without an independent test set from a separate data source(s). DL models are likely to overfit training data distribution when independent test sets are not utilized or are prone to learn dataset-specific artifacts rather than the actual disease characteristics and underlying pathology. This study aims to assess the promise of such DL methods and datasets by investigating the key challenges and issues by examining the compositions of the available public image datasets and designing different experimental setups. A convolutional neural network-based network, called CVR-Net (COVID-19 Recognition Network), has been proposed for conducting comprehensive experiments to validate our hypothesis. The presented end-to-end CVR-Net is a multi-scale-multi-encoder ensemble model that aggregates the outputs from two different encoders and their different scales to convey the final prediction probability. Three different classification tasks, such as 2-, 3-, 4-classes, are designed where the train–test datasets are from the single, multiple, and independent sources. The obtained binary classification accuracy is 99.8% for a single train–test data source, where the accuracies fall to 98.4% and 88.7% when multiple and independent train–test data sources are utilized. Similar outcomes are noticed in multi-class categorization tasks for single, multiple, and independent data sources, highlighting the challenges in developing DL models with the existing public datasets without an independent test set from a separate dataset. Such a result concludes a requirement for a better-designed dataset for developing DL tools applicable in actual clinical settings. The dataset should have an independent test set; for a single machine or hospital source, have a more balanced set of images for all the prediction classes; and have a balanced dataset from several hospitals and demography. Our source codes and model are publicly available1 for the research community for further improvements.
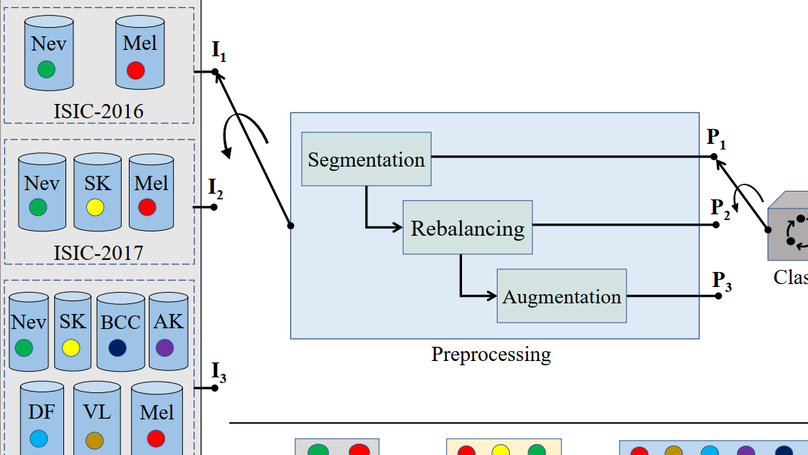
Background and Objective: Although automated Skin Lesion Classification (SLC) is a crucial integral step in computer-aided diagnosis, it remains challenging due to variability in textures, colors, indistinguishable boundaries, and shapes. Methods: This article proposes an automated dermoscopic SLC framework named Dermoscopic Expert (Dermo- Expert). It combines the pre-processing and hybrid Convolutional Neural Network (hybrid-CNN). The proposed hybrid-CNN has three distinct feature extractor modules, which are fused to achieve better-depth feature maps of the lesion. Those single and fused feature maps are classified using different fully connected layers, then ensembled to predict a lesion class. In the proposed pre-processing, we apply lesion segmentation, augmentation (geometry- and intensity-based), and class rebalancing (penalizing the majority class’s loss and merging additional images to the minority classes). Moreover, we leverage transfer learning from the pre-trained models. Finally, we deploy the weights of our DermoExpert to a possible web application. Results: We evaluate our DermoExpert on the ISIC-2016, ISIC-2017, and ISIC-2018 datasets, where the DermoExpert has achieved the area under the receiver operating characteristic curve (AUC) of 0.96, 0.95, and 0.97, respectively. The experimental results improve the state-of-the-art by the margins of 10.0% and 2.0%, respectively, for the ISIC-2016 and ISIC-2017 datasets in terms of AUC. The DermoExpert also outperforms by 3.0% for the ISIC-2018 dataset concerning a balanced accuracy. Conclusion: Since DermoExpert provides better classification outcomes on three different datasets, leading to a better recognition tool to assist dermatologists. Our source code and segmented masks for the ISIC-2018 dataset will be available as a public benchmark for future improvements.
Steady-state Visually Evoked Potential (SSVEP) based Electroencephalogram (EEG) signal is utilized in brain-computer interface paradigms, diagnosis of brain diseases, and measurement of the cognitive status of the human brain. However, various artifacts such as the Electrocardiogram (ECG), Electrooculogram (EOG), and Electromyogram (EMG) are present in the raw EEG signal, which adversely affect the EEG-based appliances. In this research, Adaptive Neuro-fuzzy Interface Systems (ANFIS) and Hilbert-Huang Transform (HHT) are primarily employed to remove the artifacts from EEG signals. This work proposes Adaptive Noise Cancellation (ANC) and ANFIS based methods for canceling EEG artifacts. A mathematical model of EEG with the aforementioned artifacts is determined to accomplish the research goal, and then those artifacts are eliminated based on their mathematical characteristics. ANC, ANFIS, and HHT algorithms are simulated on the MATLAB platform, and their performances are also justified by various error estimation criteria using hardware implementation.
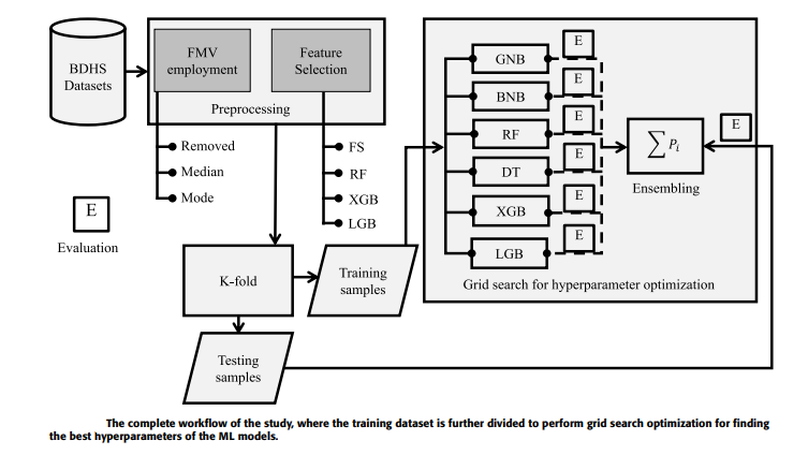
Measles is one of the significant public health issues responsible for the high mortality rate around the globe, especially in developing countries. Using nationally representative demographic and health survey data, measles vaccine utilization has been classified, and its underlying factors are identified through an ensemble Machine Learning (ML) approach. Firstly, missing values are imputed by employing various approaches, and then several feature selection techniques are applied to identify the crucial attributes for predicting measles vaccination. A grid search hyperparameter optimization technique has been applied to tune the critical hyperparameters of different ML models, such as Naive Bayes, random forest, decision tree, XGboost, and lightgbm. The individual optimized ML model’s categorization performance, as all their ensembles have been reported utilizing our proposed BDHS dataset. Individually, the optimized lightgbm provides the highest precision and AUC of 79.90% and 77.80%, respectively. This result improved when the optimized lightgbm was ensembled with XGboost, providing precision and AUC of 84.60% and 80.0%, respectively. Our result reveals that the statistical median imputation technique with the XGboost-based attribute selection method and the lightgbm classifier provides the best individual result. The performance improved when the proposed weighted ensemble of the XGboost and lightgbm approach was adopted with the same preprocessing and recommended for measles vaccine utilization. The significance of our proposed approach is that it utilizes minimum attributes collected from the child and their family members and yielded 80.0% accuracy, making it easily explainable by caregivers and healthcare personnel. Finally, our predictive model provides an early detection procedure to help national policymakers enforce new policies with specific rules and regulations.
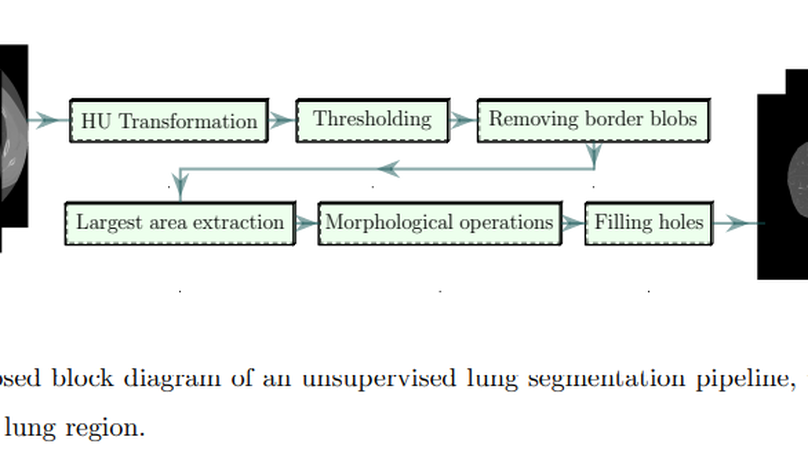
The novel COVID-19 is a global pandemic disease overgrowing worldwide. Computer-aided screening tools with greater sensitivity are imperative for disease diagnosis and prognosis as early as possible. It also can be a helpful tool in triage for testing and clinical supervision of COVID-19 patients. However, designing such an automated tool from non-invasive radiographic images is challenging as many manually annotated datasets are not publicly available yet, which is the essential core requirement of supervised learning schemes. This article proposes a 3D Convolutional Neural Network (CNN)-based classification approach considering both the inter-and intra-slice spatial voxel information. The proposed system is trained end-to-end on the 3D patches from the whole volumetric Computed Tomography (CT) images to enlarge the number of training samples, performing the ablation studies on patch size determination. We integrate progressive resizing, segmentation, augmentations, and class-rebalancing into our 3D network. The segmentation is a critical prerequisite step for COVID-19 diagnosis enabling the classifier to learn prominent lung features while excluding the outer lung regions of the CT scans. We evaluate all the extensive experiments on a publicly available dataset named MosMed, having binary- and multi-class chest CT image partitions. Our experimental results are very encouraging, yielding areas under the Receiver Operating Characteristics (ROC) curve of 0.914±0.049 and 0.893±0.035 for the binary- and multi-class tasks, respectively, applying 5-fold cross-validations. Our method’s promising results delegate it as a favorable aiding tool for clinical practitioners and radiologists to assess COVID-19.
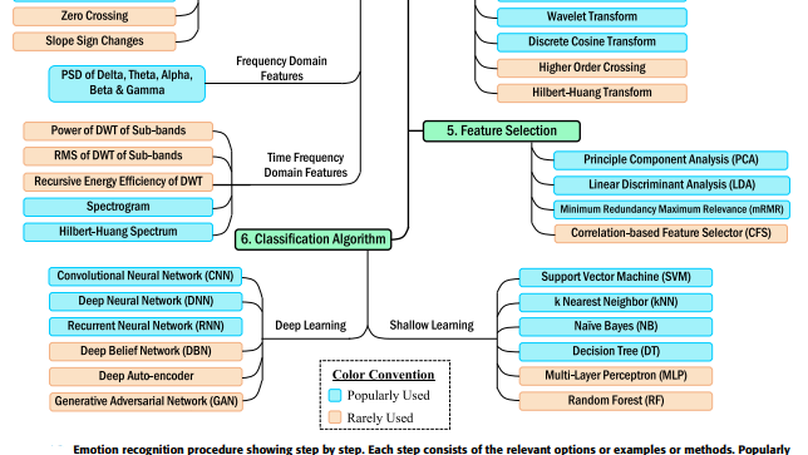
Recently, electroencephalogram-based emotion recognition has become crucial in enabling the Human-Computer Interaction (HCI) system to become more intelligent. Due to the outstanding applications of emotion recognition, e.g., person-based decision making, mind-machine interfacing, cognitive interaction, affect detection, feeling detection, etc., emotion recognition has become successful in attracting the recent hype of AI-empowered research. Therefore, numerous studies have been conducted driven by a range of approaches, which demand a systematic review of methodologies used for this task with their feature sets and techniques. It will facilitate the beginners as guidance towards composing an effective emotion recognition system. In this article, we have conducted a rigorous review on the state-of-the-art emotion recognition systems, published in recent literature, and summarized some of the common emotion recognition steps with relevant definitions, theories, and analyses to provide key knowledge to develop a proper framework. Moreover, studies included here were dichotomized based on two categories: i) deep learning-based, and ii) shallow machine learning-based emotion recognition systems. The reviewed systems were compared based on methods, classifier, the number of classified emotions, accuracy, and dataset used. An informative comparison, recent research trends, and some recommendations are also provided for future research directions.
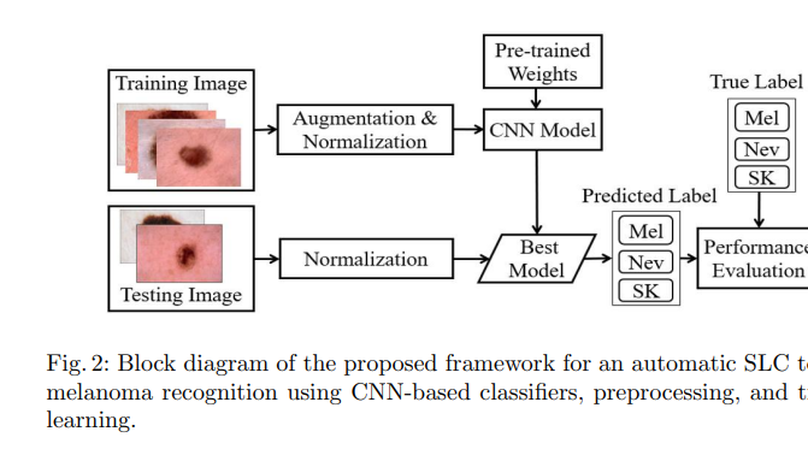
Skin cancer, also known as melanoma, is generally diagnosed visually from the dermoscopic images, which is a tedious and timeconsuming task for the dermatologist. Such a visual assessment, via the naked eye for skin cancers, is a challenging and arduous due to different artifacts such as low contrast, various noise, presence of hair, fiber, and air bubbles, etc. This article proposes a robust and automatic framework for the Skin Lesion Classification (SLC), where we have integrated image augmentation, Deep Convolutional Neural Network (DCNN), and transfer learning. The proposed framework was trained and tested on publicly available IEEE International Symposium on Biomedical Imaging (ISBI)- 2017 dataset. The obtained average area under the receiver operating characteristic curve (AUC), recall, precision, and F1-score are respectively 0.87, 0.73, 0.76, and 0.74 for the SLC. Our experimental studies for lesion classification demonstrate that the proposed approach can successfully distinguish skin cancer with a high degree of accuracy, which has the capability of skin lesion identification for melanoma recognition.
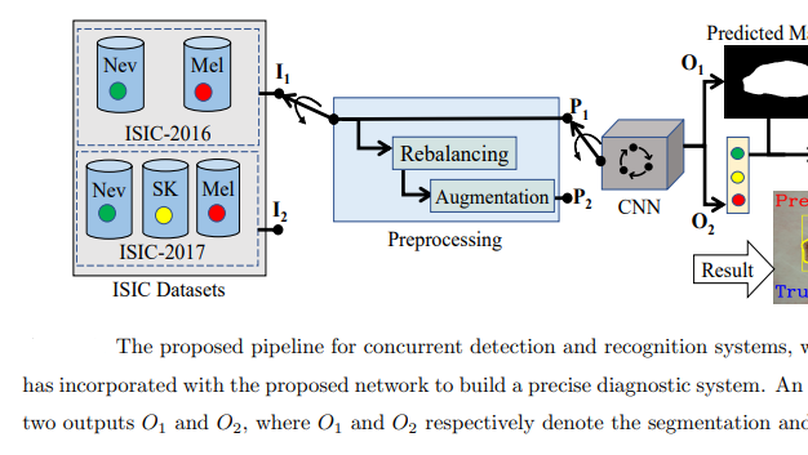
This article proposes an end-to-end deep CNN-based framework for simultaneous detection and recognition of skin lesions, Dermo-DOCTOR, consisting of two encoders. The feature maps from two encoders are fused channel-wise, called Fused Feature Map (FFM). The FFM is utilized for decoding in the detection sub-network, concatenating each stage of two encoders’ outputs with corresponding decoder layers to retrieve the lost spatial information due to pooling in the encoders. For the recognition sub-network, the outputs of three fully connected layers, utilizing feature maps of two encoders and FFM, are aggregated to obtain a final lesion class. We train and evaluate the proposed Dermo-Doctor utilizing two publicly available benchmark datasets, such as ISIC-2016 and ISIC-2017. The achieved segmentation results exhibit mean intersection over unions of 85.0% and 80.0%, respectively, for ISIC-2016 and ISIC-2017 test datasets. The proposed Dermo-DOCTOR also demonstrates praiseworthy success in lesion recognition, providing the areas under the receiver operating characteristic curves of 0.98 and 0.91 for those two datasets, respectively. The experimental results show that the proposed Dermo-DOCTOR outperforms the alternative methods mentioned in the literature for skin lesion detection and recognition. As the Dermo-DOCTOR provides better results on two different test datasets, it can be an auspicious computer-aided assistive tool for dermatologists even with limited training data.
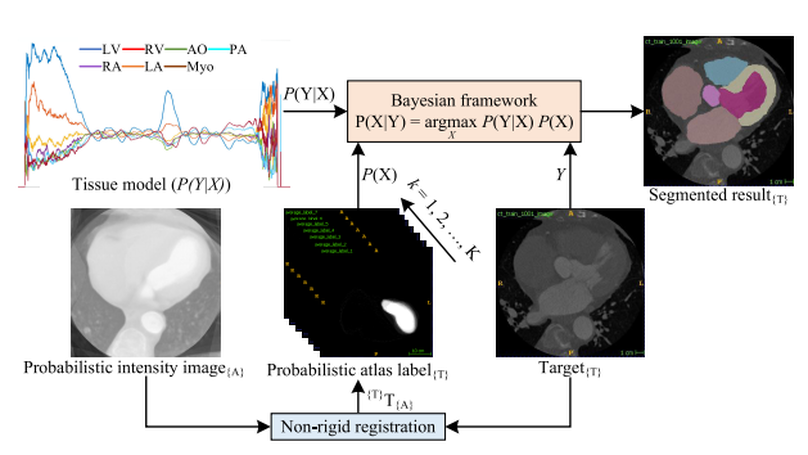
Accurate and robust whole heart substructure segmentation is crucial in developing clinical applications, such as computer-aided diagnosis and computer-aided surgery. However, the segmentation of different heart substructures is challenging because of inadequate edge or boundary information, the complexity of the background and texture, and the diversity in different substructures’ sizes and shapes. This article proposes a framework for multi-class whole heart segmentation employing non-rigid registration-based probabilistic atlas incorporating the Bayesian framework. We also propose a non-rigid registration pipeline utilizing a multi-resolution strategy for obtaining the highest attainable mutual information between the moving and fixed images. We further incorporate non-rigid registration into the expectation-maximization algorithm and implement different deep convolutional neural network-based encoder-decoder networks for ablation studies. All the extensive experiments are conducted utilizing the publicly available dataset for the whole heart segmentation containing 20 MRI and 20 CT cardiac images. The proposed approach exhibits an encouraging achievement, yielding a mean volume overlapping error of 14.5 % for CT scans exceeding the state-of-the-art results by a margin of 1.3 % in terms of the same metric. As the proposed approach provides better results to delineate the different substructures of the heart, it can be a medical diagnostic aiding tool for helping experts with quicker and more accurate results.
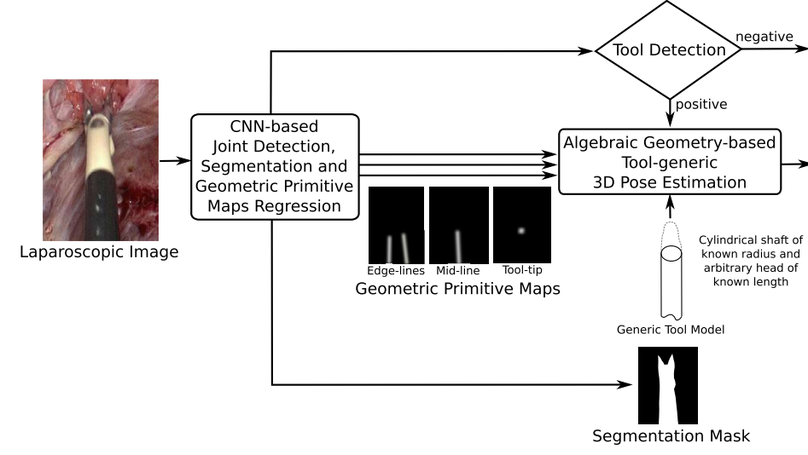
Surgical tool detection, segmentation, and 3D pose estimation are crucial components in Computer-Assisted Laparoscopy (CAL). The existing frameworks have two main limitations. First, they do not integrate all three components. Integration is critical; for instance, one should not attempt computing pose if detection is negative. Second, they have particular requirements, such as the availability of a CAD model. We propose an integrated and generic framework whose sole requirement for the 3D pose is that the tool shaft is cylindrical. Our framework makes the most of deep learning and geometric 3D vision by combining a proposed Convolutional Neural Network (CNN) with algebraic geometry. We show two applications of our framework in CAL: tool-aware rendering in Augmented Reality (AR) and tool-based 3D measurement. We name our CNN as ART-Net (Augmented Reality Tool Network). It has a Single Input Multiple Output (SIMO) architecture with one encoder and multiple decoders to achieve detection, segmentation, and geometric primitive extraction. These primitives are the tool edge-lines, mid-line, and tip. They allow the tool’s 3D pose to be estimated by a fast algebraic procedure. The framework only proceeds if a tool is detected. The accuracy of segmentation and geometric primitive extraction is boosted by a new Full-resolution feature map Generator (FrG). We extensively evaluate the proposed framework with the EndoVis and new proposed datasets. We compare the segmentation results against several Fully Convolutional Networks (FCN) and U-Net variants. Several ablation studies are provided for detection, segmentation, and geometric primitive extraction. The proposed datasets are surgery videos of different patients. In detection, ART-Net achieves 100.0% in both average precision and accuracy. In segmentation it achieves 81.0% in mean Intersection over Union (mIoU) on the robotic EndoVis dataset (articulated tool), where it outperforms both FCN and UNet by 4.5pp and 2.9pp, respectively. It achieves 88.2% mIoU on the remaining datasets (non-articulated tool). In geometric primitive extraction, ART-Net achieves 2.45 degrees and 2.23 degrees in mean Arc Length (mAL) error for the edge lines and mid-line, respectively, and 9.3 pixels in mean Euclidean distance error for the tooltip. Finally, regarding the 3D pose evaluated on animal data, our framework achieves 1.87 mm, 0.70 mm, and 4.80 mm mean absolute errors on the X, Y, and Z coordinates, respectively, and 5.94∘ angular errors on the shaft orientation. It achieves 2.59 mm and 1.99 mm in the tool head’s mean and median location error evaluated on patient data. The proposed framework outperforms existing ones in detection and segmentation. Compared to separate networks, integrating the tasks in a single network preserves accuracy in detection and segmentation but substantially improves accuracy in geometric primitive extraction. Our framework has similar or better accuracy in 3D pose estimation while improving robustness against laparoscopy’s very challenging imaging conditions.
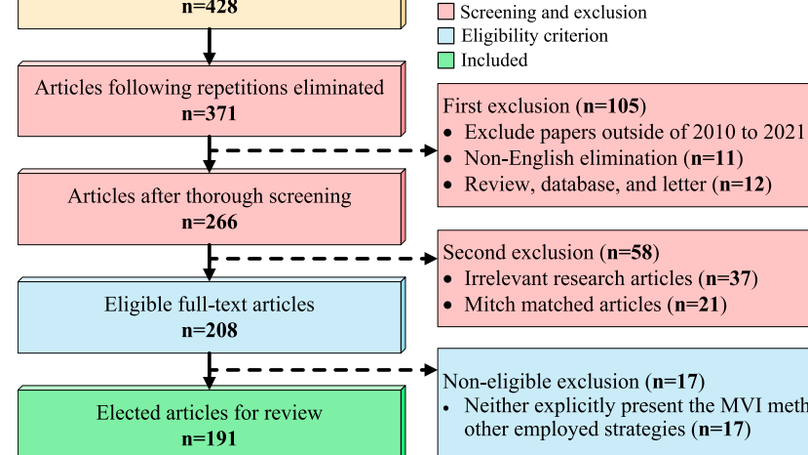
Recently, numerous studies have been conducted on Missing Value Imputation (MVI), intending the primary solution scheme for the datasets containing one or more missing attribute’s values. The incorporation of MVI reinforces the Machine Learning (ML) models’ performance and necessitates a systematic review of MVI methodologies employed for different tasks and datasets. It will aid beginners as guidance towards composing an effective ML-based decision-making system in various fields of applications. This article aims to conduct a rigorous review and analysis of the state-of-the-art MVI methods in the literature published in the last decade. Altogether, 191 articles, published from 2010 to August 2021, are selected for review using the well-known Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) technique. We summarize those articles with relevant definitions, theories, and analyses to provide essential information for building a precise decision-making framework. In addition, the evaluation metrics employed for MVI methods and ML-based classification models are also discussed and explored. Remarkably, the trends for the MVI method and its evaluation are also scrutinized from the last twelve years’ data. To come up with the conclusion, several ML-based pipelines, where the MVI schemes are incorporated for performance enhancement, are investigated and reviewed for many different datasets. In the end, informative observations and recommendations are addressed for future research directions and trends in related fields of interest.
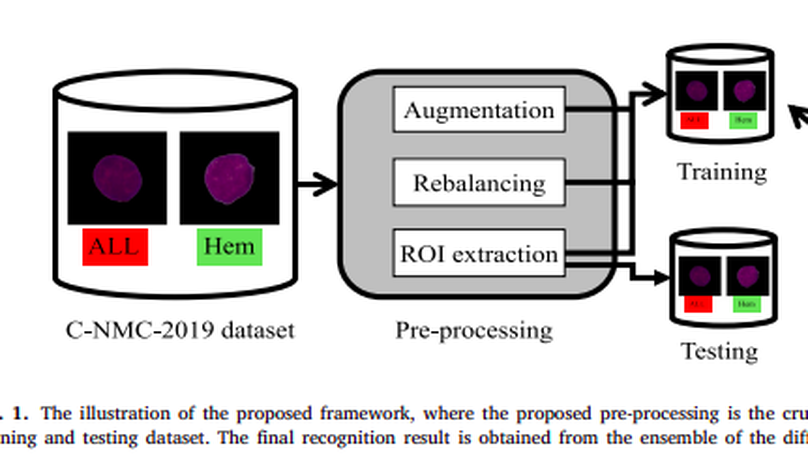
Acute Lymphoblastic Leukemia (ALL) is a blood cell cancer characterized by the presence of excess immature lymphocytes., Even though automation in ALL prognosis is essential for cancer diagnosis, it remains a challenge due to the morphological correlation between malignant and normal cells. The traditional ALL classification strategy demands that experienced pathologists read cell images carefully, which is arduous, time-consuming, and often hampered by interobserver variation. This article has automated the ALL recognition task by employing deep Convolutional Neural Networks (CNNs). The weighted ensemble of deep CNNs is explored to recommend a better ALL cell classifier. The weights are estimated from ensemble candidates’ corresponding metrics, such as F1-score, area under the curve (AUC), and kappa values. Various data augmentations and pre-processing are incorporated to achieve a better generalization of the network. Our proposed model was trained and evaluated utilizing the C-NMC-2019 ALL dataset. The proposed weighted ensemble model has outputted a weighted F1-score of 89.7%, a balanced accuracy of 88.3%, and an AUC of 0.948 in the preliminary test set. The qualitative results displaying the gradient class activation maps confirm that the introduced model has a concentrated learned region. In contrast, the ensemble candidate models, such as Xception, VGG-16, DenseNet-121, MobileNet, and InceptionResNet-V2, separately exhibit coarse and scatter learned areas in most cases. Since the proposed ensemble yields a better result for the aimed task, it can support clinical decisions to detect ALL patients in an early stage.
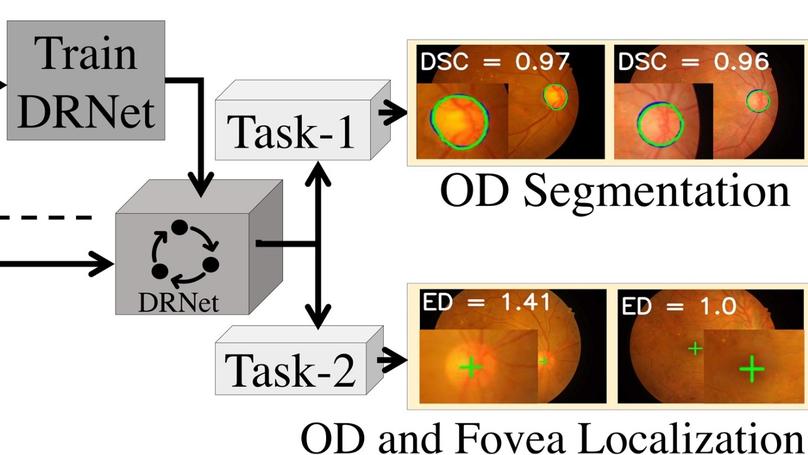
This article introduces a novel end-to-end encoder-decoder network, DRNet, explicitly designed for the segmentation and localization of OD and Fovea centers. In our DRNet, we propose a skip connection, named residual skip connection, to compensate for the spatial information lost due to pooling in the encoder. Unlike the earlier skip connection in the UNet, the proposed skip connection does not directly concatenate low-level feature maps from the encoder’s beginning layers with the corresponding same-scale decoder. We validate DRNet using different publicly available datasets, such as IDRiD, RIMONE, DRISHTI-GS, and DRIVE for OD segmentation; IDRiD and HRF for OD center localization; and IDRiD for Fovea center localization. The proposed DRNet, for OD segmentation, achieves mean Intersection over Union (mIoU) of 0.845, 0.901, 0.933, and 0.920 for IDRiD, RIMONE, DRISHTI-GS, and DRIVE, respectively. Our OD segmentation result, in terms of mIoU, outperforms the state-of-the-art results for IDRiD and DRIVE datasets, whereas it outperforms state-of-the-art results concerning mean sensitivity for RIMONE and DRISHTI-GS datasets. The DRNet localizes the OD center with mean Euclidean Distance (mED) of 20.23 and 13.34 pixels, respectively, for IDRiD and HRF datasets; it outperforms the state-of-the-art by 4.62 pixels for IDRiD dataset. The DRNet also successfully localizes the Fovea center with mED of 41.87 pixels for the IDRiD dataset, outperforming the state-of-the-art by 1.59 pixels for the same dataset. The proposed DRNet, for OD segmentation, achieves mean Intersection over Union (mIoU) of 0.845, 0.901, 0.933, and 0.920 for IDRiD, RIMONE, DRISHTI-GS and DRIVE, respectively. Our OD segmentation result, in terms of mIoU, outperforms the state-of-the-art results for IDRiD and DRIVE datasets, whereas it outperforms state-of-the-art results concerning mean sensitivity for RIMONE and DRISHTI-GS datasets. The DRNet localizes the OD center with mean Euclidean Distance (mED) of 20.23 and 13.34 pixels, respectively, for IDRiD and HRF datasets; it outperforms the state-of-the-art by 4.62 pixels for IDRiD dataset. The DRNet also successfully localizes the Fovea center with mED of 41.87 pixels for the IDRiD dataset, outperforming the state-of-the-art by 1.59 pixels for the same dataset.
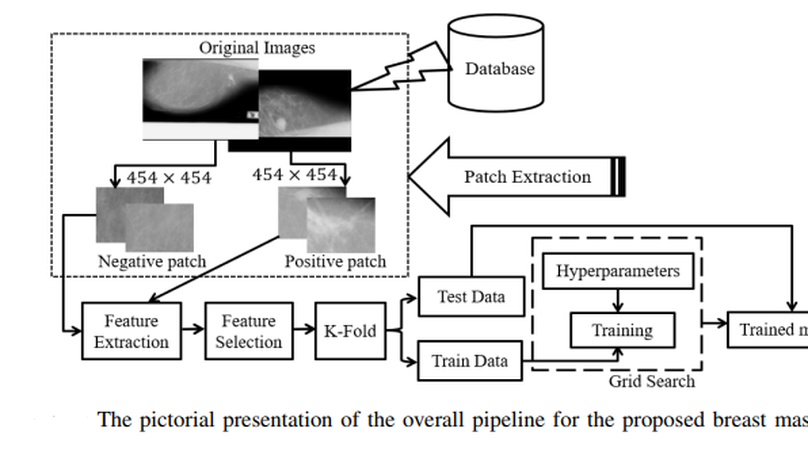
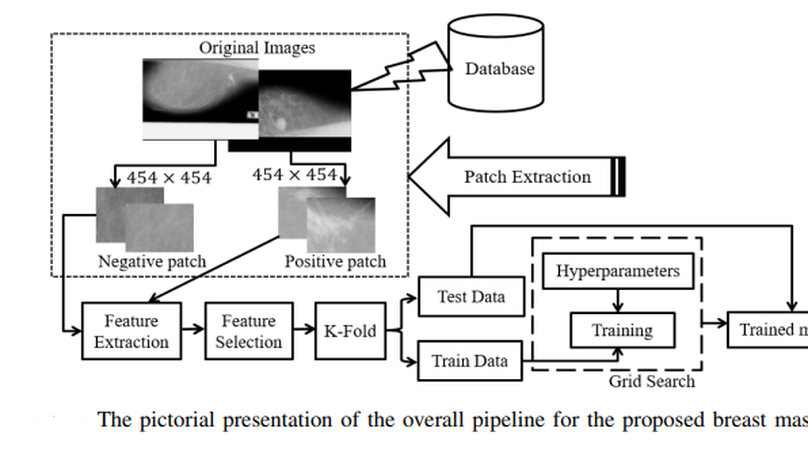
Mammography is the most widely used gold standard for screening breast cancer, where mass classification is a prominent step. Classification of mass in the breast is, however, an arduous problem as they usually have large variations in terms of shape, size, boundary, and texture. In this study, the process of mass classification is automated with the use of transfer learning of Deep Convolutional Neural Networks (DCNN) to extract features, the bagged decision tree for feature selection, and finally a Support Vector Machine (SVM) classifier for classifying the mass and non-mass tissue. Area Under ROC Curve (AUC) is chosen as the performance metric, which is then maximized for hyper-parameter tuning using a grid search. All experiments, in this paper, were conducted using the INbreast dataset. The best obtained AUC from the experimental results is O.994±0.003. Our results conclude that high-level distinctive features can be extracted from Mammograms by using the pre-trained DCNN, which can be used with the SVM classifier to robustly distinguish between the mass and non-mass presence in the breast.
The risk factors and severity of diabetes can be reduced significantly if a precise early prediction is possible. The robust and accurate prediction of diabetes is highly challenging due to the limited number of labeled data and also the presence of outliers (or missing values) in the diabetes datasets. This paper proposes a robust framework for diabetes prediction where the outlier rejection, filling the missing values, data standardization, feature selection, K-fold cross-validation, and different machine learning (ML) classifiers (k-nearest Neighbour, Decision Trees, Random Forest, AdaBoost, Naive Bayes, and XGBoost) and multilayer perceptron (MLP) were employed. The weighted ensembling of different ML models is also proposed to improve diabetes prediction, where the weights are estimated from the corresponding area under the ROC curve (AUC) of the ML model. AUC is chosen as the performance metric, which is then maximized during hyperparameter tuning using the grid search technique. All the experiments were conducted under the same conditions using the Pima Indian Diabetes Dataset. From all the extensive experiments, our proposed ensembling classifier is the best-performing classifier with the sensitivity, specificity, false omission rate, diagnostic odds ratio, and AUC of 0.789, 0.934, 0.092, 66.234, and 0.950, respectively, which outperforms the state-of-the-art results by 2.00 % in AUC. Our proposed framework for diabetes prediction outperforms the other methods discussed in the article. It can also provide better results on the same dataset, leading to better performance in diabetes prediction. Our source code for diabetes prediction is made publicly available.
Automatic segmentation of skin lesions is considered a crucial step in Computer-aided Diagnosis (CAD) systems for melanoma detection. Despite its significance, skin lesion segmentation remains an unsolved challenge due to their variability in color, texture, and shape and indistinguishable boundaries. Through this study, we present a new and automatic semantic segmentation network for robust skin lesion segmentation named Dermoscopic Skin Network (DSNet). In order to reduce the number of parameters to make the network lightweight, we used a depth-wise separable convolution instead of standard convolution to project the learned discriminating features onto the pixel space at different stages of the encoder. We also implemented a U-Net and a Fully Convolutional Network (FCN8s) to compare against the proposed DSNet. We evaluate our proposed model on two publicly available datasets, ISIC-2017 and PH2. The obtained mean Intersection over Union (mIoU) is 77.5% and 87.0%, respectively, for ISIC-2017 and PH2 datasets, which outperformed the ISIC-2017 challenge winner by 1.0% concerning mIoU. Our proposed network outperformed U-Net and FCN8s by 3.6% and 6.8% concerning mIoU on the ISIC-2017 dataset, respectively. Our network for skin lesion segmentation outperforms the other methods discussed in the article. It can provide better-segmented masks on two different test datasets, leading to better performance in melanoma detection. Our trained model, source code, and predicted masks are made publicly available.
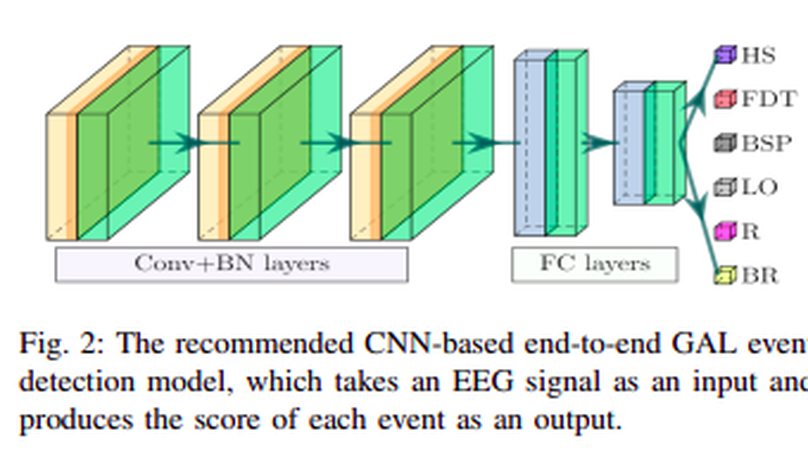
People undergoing neuromuscular dysfunctions and amputated limbs require automatic prosthetic appliances. In developing such prostheses, the precise detection of brain motor actions is imperative for the Grasp-and-Lift (GAL) tasks. Because of the low-cost and non-invasive essence of Electroencephalogra-phy (EEG), it is widely preferred for detecting motor actions while controlling prosthetic tools. This article has automated the hand movement activity viz GAL detection method from the 32-channel EEG signals. The proposed pipeline essentially combines preprocessing and end-to-end detection steps, eliminating the requirement of hand-crafted feature engineering. Preprocessing action consists of raw signal denoising, using either Discrete Wavelet Transform (DWT) or highpass or bandpass filtering and data standardization. The detection step consists of Convolutional Neural Network (CNN)- or Long Short Term Memory (LSTM)-based model. All the investigations utilize the publicly available WAY-EEG-GAL dataset, having six different GAL events. The best experiment reveals that the proposed framework achieves an average area under the ROC curve of 0.944, employing the DWT-based denoising filter, data standardization, and CNN-based detection model. The obtained outcome designates an excellent achievement of the introduced method in detecting GAL events from the EEG signals, turning it applicable to prosthetic appliances, brain-computer interfaces, robotic arms, etc.
Automatic segmentation of brain magnetic resonance imaging (MRI) images is one of the vital steps for quantitative analysis of the brain for further inspection. In this paper, NeuroNet has been adopted to segment the brain tissues using a ResNet in the encoder and a fully convolution network (FCN) in the decoder. Various hyper-parameters have been tuned to achieve the best performance, while network parameters (kernel and bias) were initialized using the NeuroNet pre-trained model. Different pre-processing pipelines have also been introduced to get a robust trained model. The model has been trained and tested on the IBSR18 data set. The outcome of the research indicates that for the IBSR18 data set, pre-processing and proper tuning of hyper-parameters for the NeuroNet model have improved DICE for brain tissue segmentation.
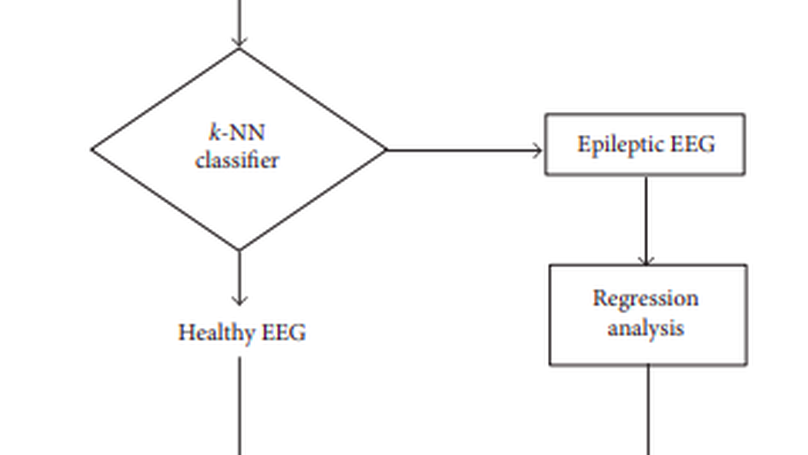
Electroencephalographic signal is a representative signal that contains information about brain activity, which is used for the detection of epilepsy since epileptic seizures are caused by a disturbance in the electrophysiological activity of the brain. The prediction of epileptic seizure usually requires a detailed and experienced analysis of EEG. In this paper, we have introduced a statistical analysis of EEG signal that is capable of recognizing epileptic seizure with a high degree of accuracy and helps to provide automatic detection of epileptic seizure for different ages of epilepsy. To accomplish the target research, we extract various epileptic features namely approximate entropy (ApEn), standard deviation (SD), standard error (SE), modified mean absolute value (MMAV), roll-off (), and zero crossing (ZC) from the epileptic signal. The -nearest neighbours (-NN) algorithm is used for the classification of epilepsy then regression analysis is used for the prediction of the epilepsy level at different ages of the patients. Using the statistical parameters and regression analysis, a prototype mathematical model is proposed which helps to find the epileptic randomness with respect to the age of different subjects. The accuracy of this prototype equation depends on proper analysis of the dynamic information from the epileptic EEG.
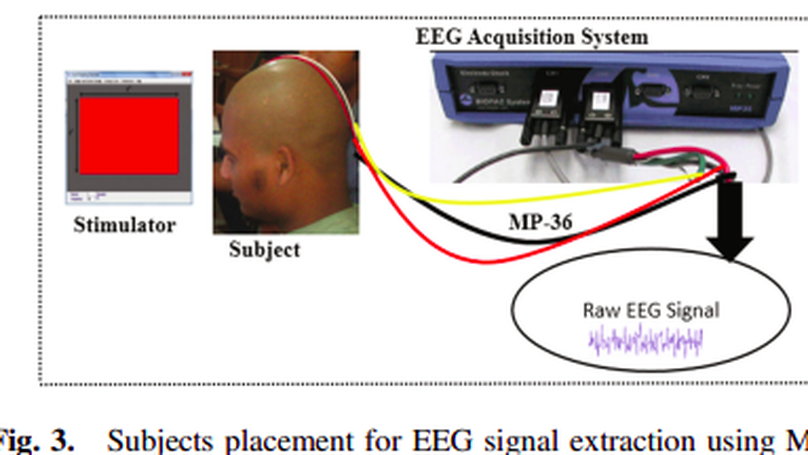
BCIs, which elaborated as Brain-computer Interface that use brain responses to control the BCI paradigms. These brain responses are measured using Electroencephalographic signal along the scalp of the subjects. However, the less variability of EEG signal from the subjects make the BCI paradigms user independent. In this research, we simply analyze the user independency of SSVEP based EEG signal that makes a conclusion inter subject’s variability of BCI users. To accomplish the research goal, SSVEP based EEG signal extract from both different subjects and different stimulation conditions and a features vector is formed to compare each subject’s variability. Artificial Neural Network classifier is used to determine the deviation and regression of deviation of each features vectors. From the heatmap and classifier, it is found that the used independency of the EEG signal is less that means that less variability of EEG. That ensures the user independent BCI paradigms with high transfer rate of the bits.
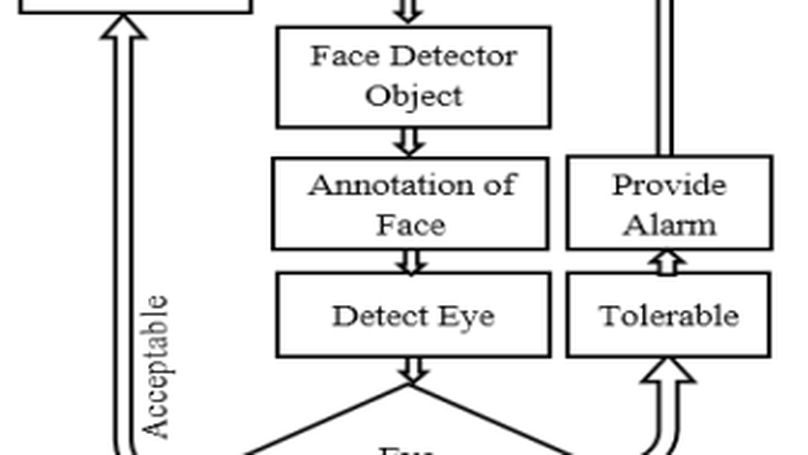
Security and reconnaissance applications are prominent BCI paradigms which are less complex and sophisticated if there is no contamination in Electroencephalogram (EEG) signal. The better the quality of EEG signal ensures the better the performance (better Information Transfer Rate (ITR), high Signal to Noise Ratio (SNR), high Bandwidth (BW), and so on) of BCI paradigms. Drowsiness is one of the major contamination in EEG signal that hampers the operation of modern BCI paradigms. In this research, a non-intrusive machine vision based concept is used to determine the drowsiness from the patient which ensure the drowsy free EEG signal. In this proposed system, a camera which placed in a way that it records subjects (BCI Users) eye movement in every time as well as it can monitor the open and close state of eye. Viola-jones Algorithm is applicable for the detection of face as well as state of eye (Open, closed or semi-open) which is the key concern for the detection of drowsiness from the patient’s EEG signal. After detecting this drowsiness, decision can be easily made for the perfect operation of BCI.
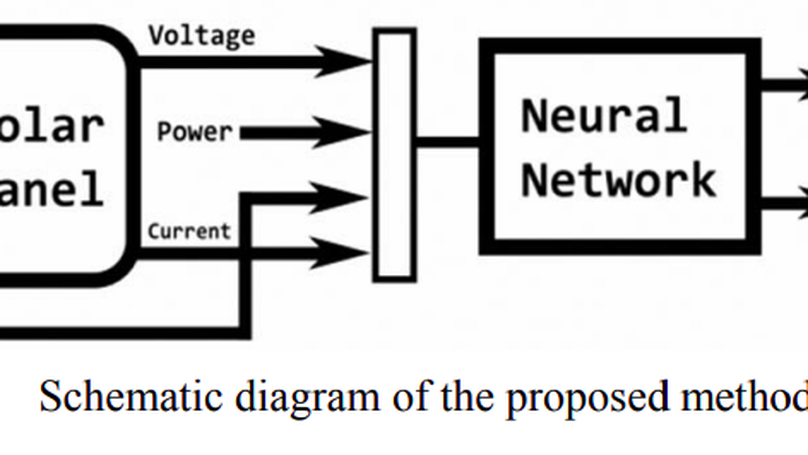
Now a days in power generation, renewable energy plays a vital role in which photovoltaic energy generation placed top in the list of the renewable energy because of the easy process of generation. The photovoltaic energy depends on the solar irradiance and the temperature. To get the maximum power from the PV panel, the idea of Maximum Power Point Tracking (MPPT) is arrived. Too many algorithms and controllers have been considered in the past to track the maximum power and to reduce the tracking time and also to improve the efficiency of PV panel. In this paper, Artificial Neural Network (ANN) techniques is proposed to track the maximum power. The proposed method has been evaluated by simulation in MATLAB environment. The simulation results show the effectiveness of the proposed technique and its ability to track the maximum power of the PV panel.
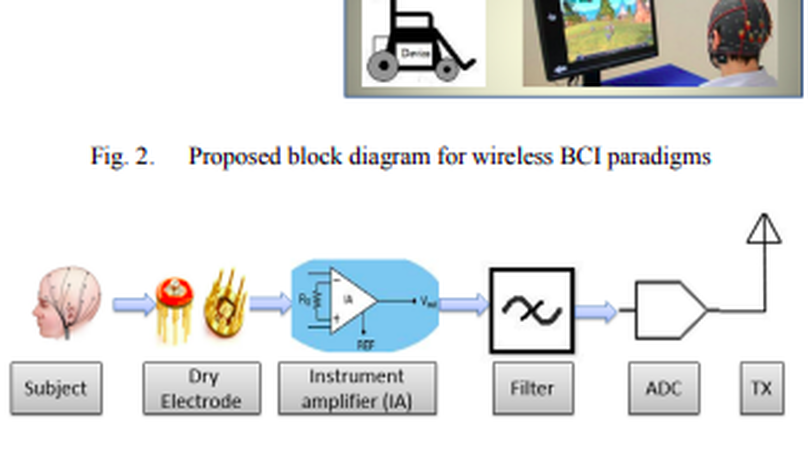
A Brain-computer Interface (BCI) is a communication pathway to provide ease to the users for interacting with the outside surroundings after translating brain signals into machine commands. The modern Steady-state Visual Evoked Potential (SSVEP) based Electroencephalographic (EEG) signals has become the most sophisticated methodology for a BCI paradigms. So, the perfection of SSVEP signal make the perfection of the BCI paradigm. The use of gel based wet electrode for the extraction of EEG signal is too much noisy and unpredictable for long time measurement which degrades the quality of SSVEP signal in a consequence degrades the performance of modern BCI paradigm. In our research, we are trying to solve this degradation of the quality of SSVEP signal. To accomplish this goal, a typical wireless BCIs using dry electrode is proposed for long term application without sacrificing Information Transfer Rate (ITR), Signal to Noise Ratio (SNR). After extracting SSVEP signal using dry electrode, Analog to Digital Conversion (ADC) is proceeded for the wireless transmission for remote BCI paradigms. Finally, after receiving this signal any BCI paradigms can be operated with high degree of accuracy.
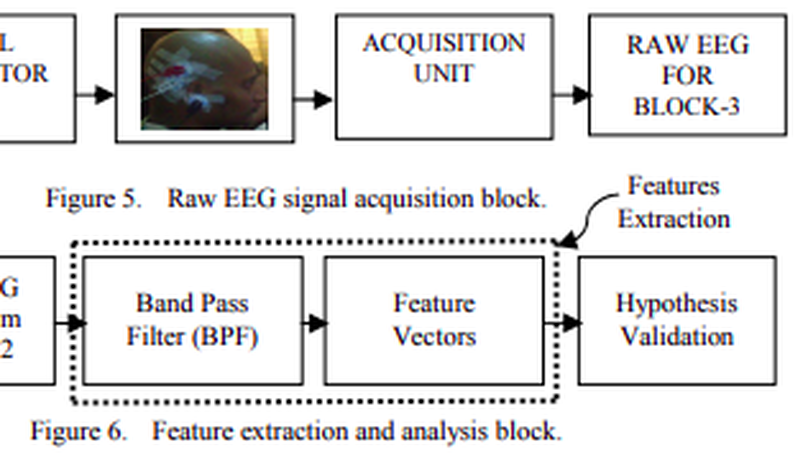
Brain-computer Interfaces (BCIs) are the communicating bridges between the human brain and a computer which may be implemented on the basis of Steady-state Visual Evoked Potentials (SSVEPs). It is mandatory to improve the stimulation for the betterment of the accuracy of modern BCIs, higher Information Transfer Rate (ITR), desired bandwidth (BW), and Signal to Noise Ratio (SNR) of BCIs. The performance of stimulator depends on many factors such as size and shape of stimulator, frequency of stimulation, luminance, color, and subject attention. Information Transfer Rate (ITR) varies with the change of frequency and size of the visual stimuli. In our research, a Circular Repetitive Visual Stimulator (CRVS) of different diameters (2″, 2.5″ and 3″), colors (RGB), frequencies (10, 15 and 20 Hz) was used. The raw EEG signal is processed for finding the effect of diverse stimulation on alpha band of EEG signal at diverse condition. From the analysis it is found that, when the size of the stimulator changes from 2″ to 2.5″, resultant increase in alpha wave is 58.18%. But for a further increase in size from 2.5″ to 3″, there is a resultant decrease in alpha wave of 45%. Similar result is found for the changes in frequencies and colors.
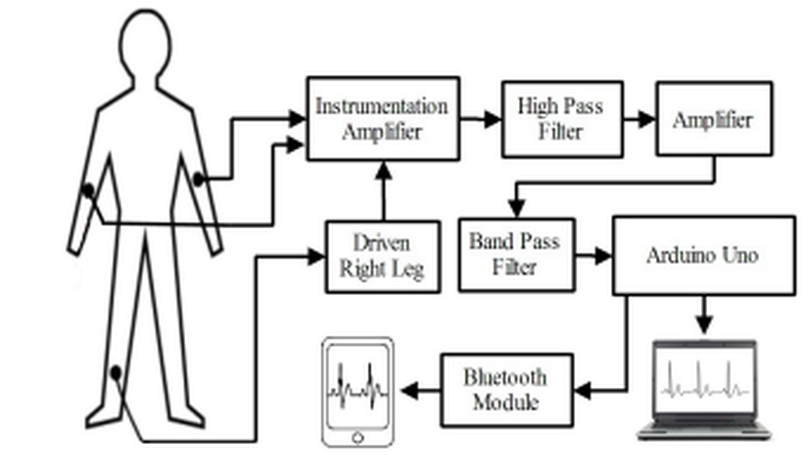
Electrocardiographic (ECG) equipment plays a vital role for diagnosis of cardiac disease. However, the cost of this equipment is huge and the operation is too much complex which cannot offer better services to a large population in developing countries like Bangladesh. In this paper, we have designed and implemented a low cost portable single channel ECG monitoring system using smartphone having android operating system and Arduino. This manuscript also demonstrates the use of Android smartphone for processing and visualizing ECG signal. Our designed system is battery powered and it gives wireless feature. This system can also be used with desktop computer or laptop having either Windows, Linux or Mac OS. For this purpose a software is developed. An Android application is developed using Processing IDE, which requires Android version 2.3 and API level of 10. This application does not need USB host API. For this reason, around 98% Android smartphone in the market can be used for this system.
ANN has been proved as a powerful discriminating classifier for tasks in medical diagnosis for early detection of diseases. In our research, ANN has been used for predicting three different diseases (heart disease, liver disorder, lung cancer). Feed-forward back propagation neural network algorithm with Multi-Layer Perceptron is used as a classifier to distinguish between infected or non-infected person. The results of applying the ANNs methodology to diagnosis of these disease based upon selected symptoms show abilities of the network to learn the patterns corresponding to symptoms of the person. In our proposed work, Multi-Layer Perceptron with having 2 hidden layer is used to predict medical diseases. Here in case of liver disorder prediction patients are classified into four categories: normal condition, abnormal condition (initial), abnormal condition and severe condition. This neural network model shows good performance in predicting disease with less error.
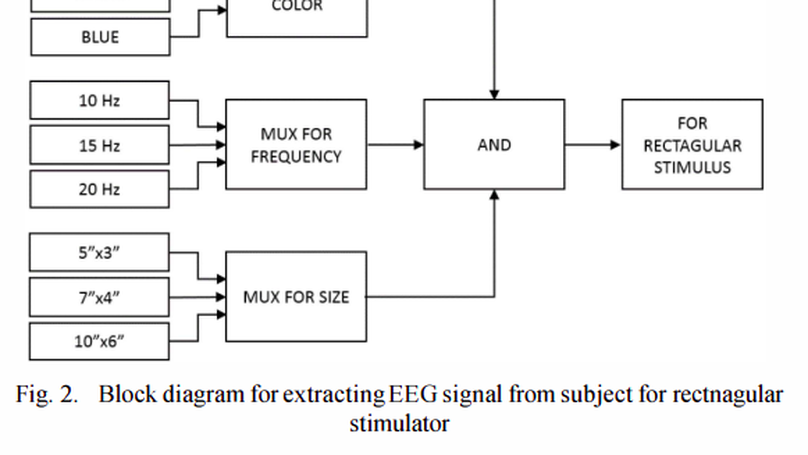
Steady-state visual evoked potential (SSVEP) is used for Brain-Computer Interface that requires little training for user, offers high information transfer rate and higher accuracy in living environments. To elicit an SSVEP, a Repetitive Visual Stimulus (RVS) of rectangular shape has been proposed to the subjects in different ways. RVS is rendered on a computer screen by flashing graphical patterns. The properties of SSVEP depend on the rendering device and frequency, size, shape and color of the stimuli. But, literatures on SSVEP-based BCI are seldom provided with reasons for usefulness of rendering devices or RVS properties. The aim of this research is to study the effect of all these stimulation properties on performance of SSVEP while elicited by a rectangular shape. A correlation matrix is made to help selection of any suitable SSVEP stimulator. The percentage change in energy and power from one kind of stimulation to another is also shown in this paper. Performance achieved in different cases has been compared with each other for apposite understanding. This will help a researcher to select proper stimulation types to elicit SSVEP.
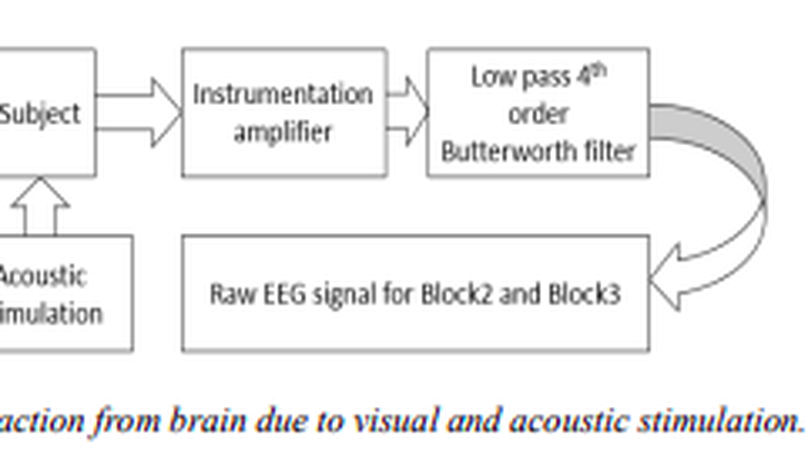
In this work the electrical activity in brain or known as electroencephalogram (EEG) signal is being analyzed to study the various effects of sound on the human brain activity. The effect is in the form of variation in either frequency or in the power of different EEG bands. A biological EEG signal stimulated by Music listening reflects the state of mind, impacts the analytical brain and the subjective-artistic brain. A two channel EEG acquisition unit is being used to extract brain signal with high transfer rate as well as good SNR. This paper focused on three types of brain waves which are theta (4-7 Hz), alpha (8-12 Hz) and beta wave (13-30 Hz). The analysis is carried out using Power Spectral density (PSD), Correlation co-efficient analysis. The outcome of this research depicted that high amplitude Alpha and low amplitude Beta wave and low amplitude Alpha and high amplitude Beta wave is associated with melody and rock music respectively meanwhile theta has no effect. High power of alpha waves and low power of beta waves that obtained during low levels of sound (Melody) indicate that subjects were in relaxed state. When subjects exposed to high level of sound (Rock), beta waves power increased indicating subjects in disturbed state. Meanwhile, the decrease of alpha wave magnitude showed that subjects in tense. Thus the subject’s executional attention level is determined by analyzing the different components of EEG signal.
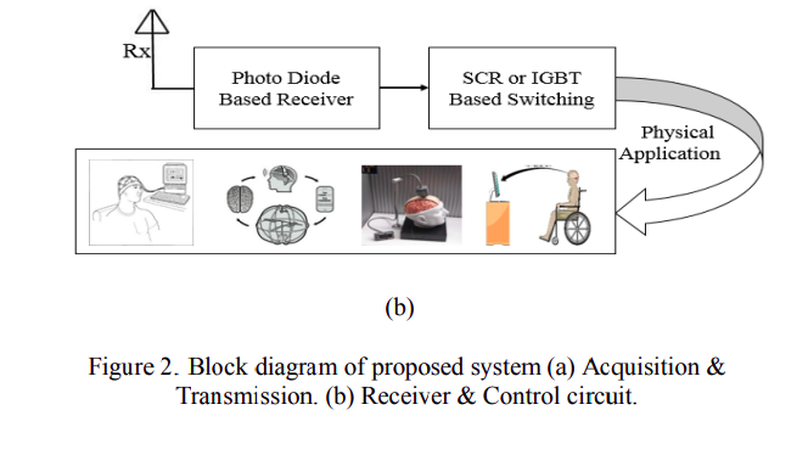
The motive of this paper is to design a low cost wireless EEG acquisition system for easily monitoring of the patient. Using local effort and low price employment, this system can be built which includes data acquisition, data transmission, and receiving unit which contains the patient monitoring site. The developed wireless EEG system is also suitable for the applications such as remote control of devices, rescue, etc. Realtime decoding and mobile EEG signal processing with high information transfer rate (ITR) are incorporated in the system. The specialty of the proposed research is inclusion of forth order Butterworth low pass filter which has better stability and sharper cut off with reasonable cost. Using this techniques hardware implementation is possible and GSM system can be added with hardware for long distance wireless transmission of EEG signal. The system performance is simulated by some simulation software. The proposed system is reliable, and cost is about 950 BDT or 12 USD which is reasonable.
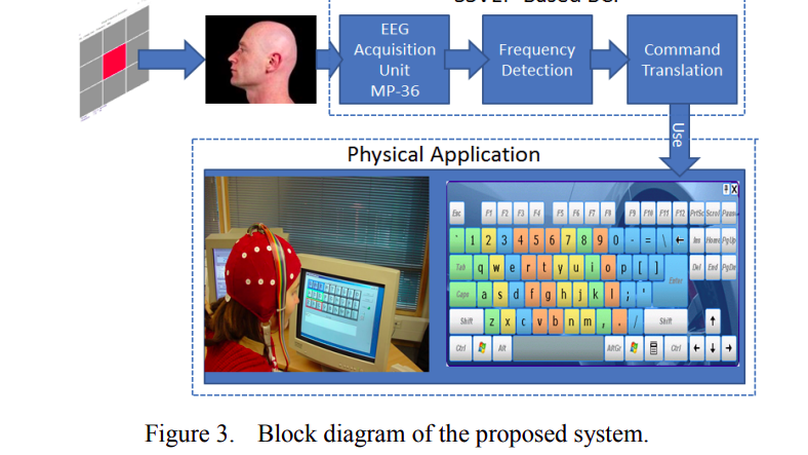
A Brain Computer Interface (BCI) provides a new communication channel between human brain and the computer. This paper presents a direct noninvasive brain-computer interface (BCI) that can help users to select any command in the graphical user interface (GUI) such as appliance control, cursor control, typing, making phone call, etc. The system is based on the steady-state visual evoked potential (SSVEP) which use three fixed positioned electrodes for reducing user variation on system performance. The frequency-coded SSVEP is used with different stimuli procedure such as effect of color and angular position of the screen which make the system user independent. The average transfer rate over all subjects is found up to 66.81 bits/min. The attractive features of the system are noninvasive signal recording, little training required for user, higher information transfer rate (HITR) and higher accuracy in living environments.